Functional Genomics
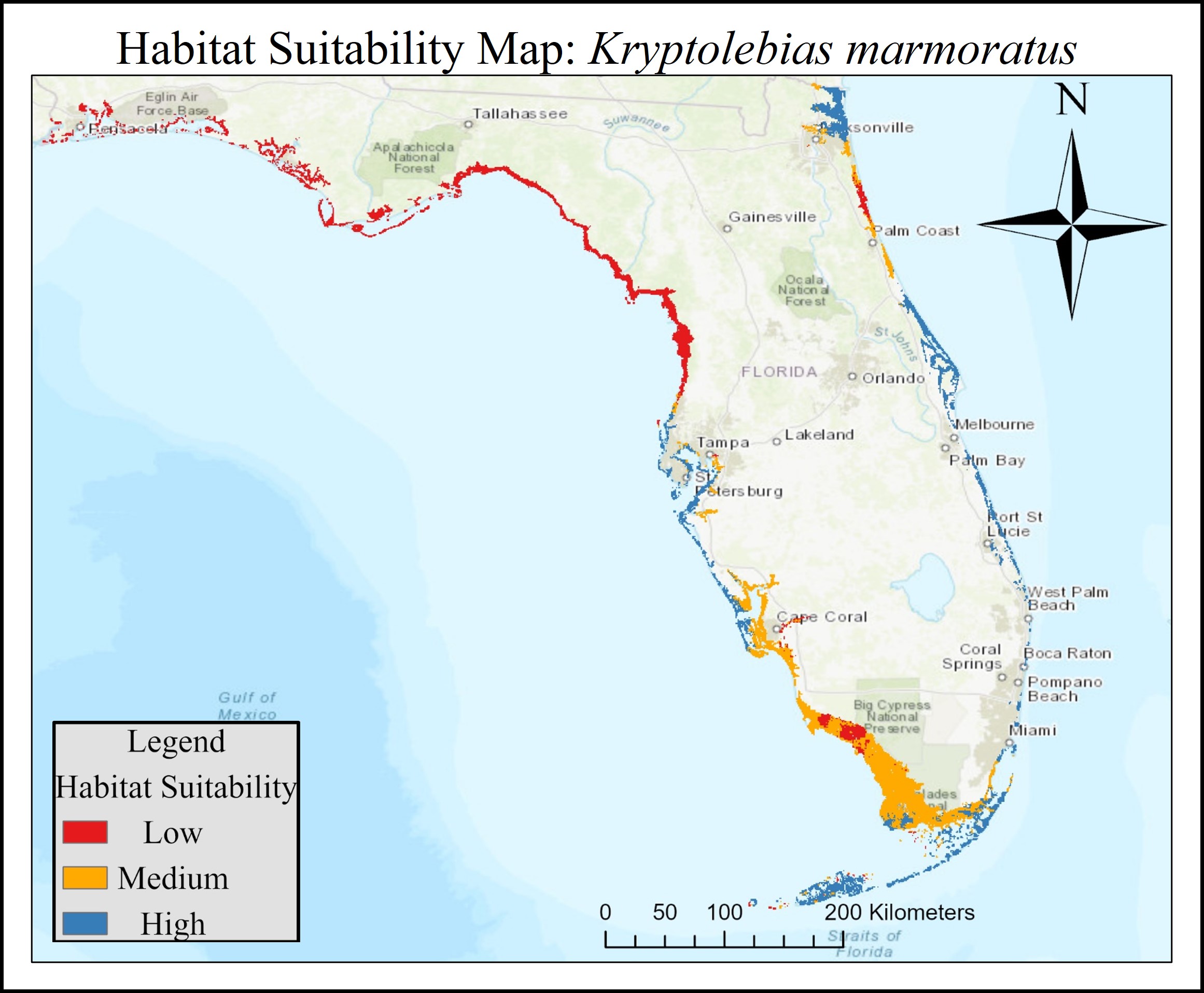
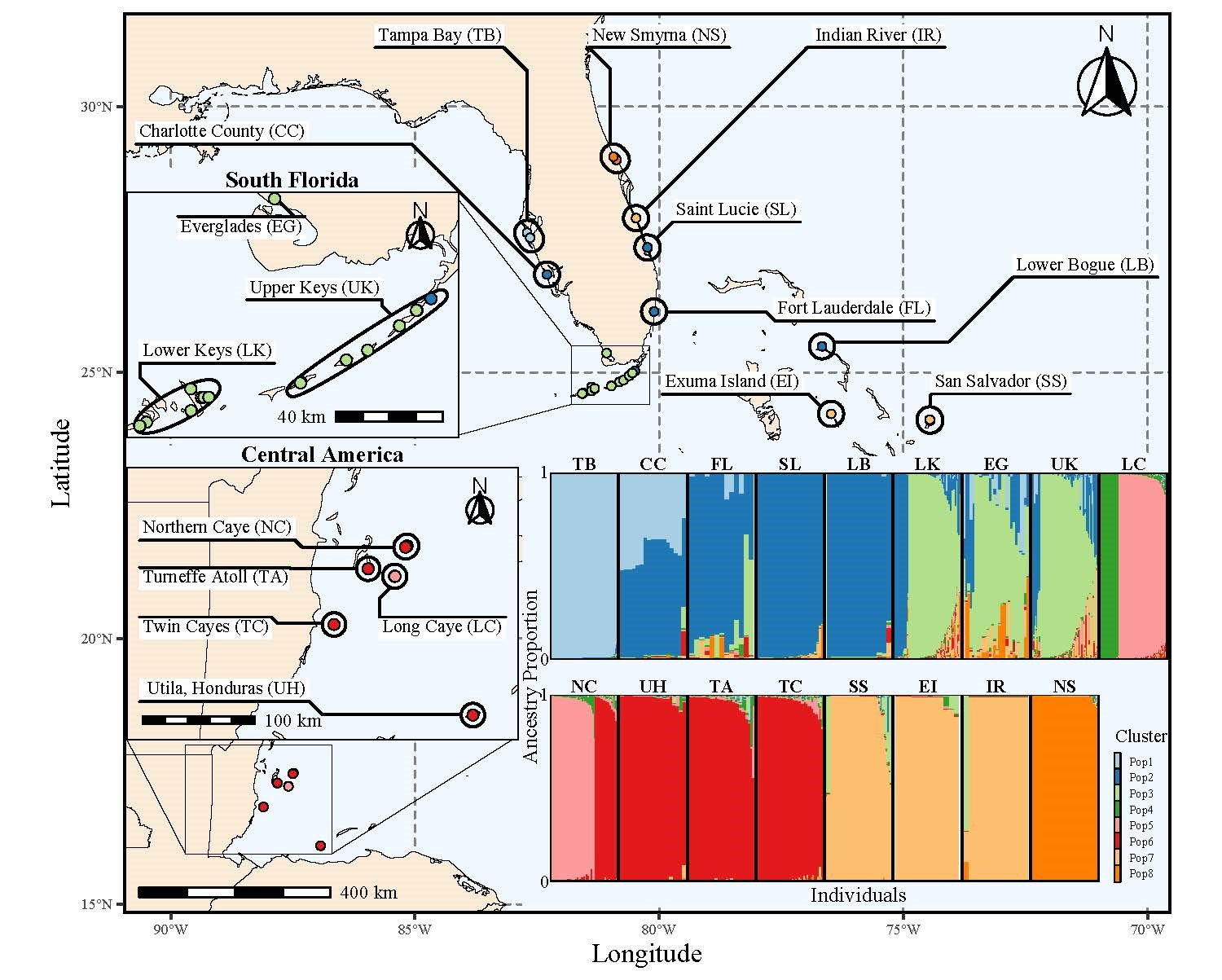
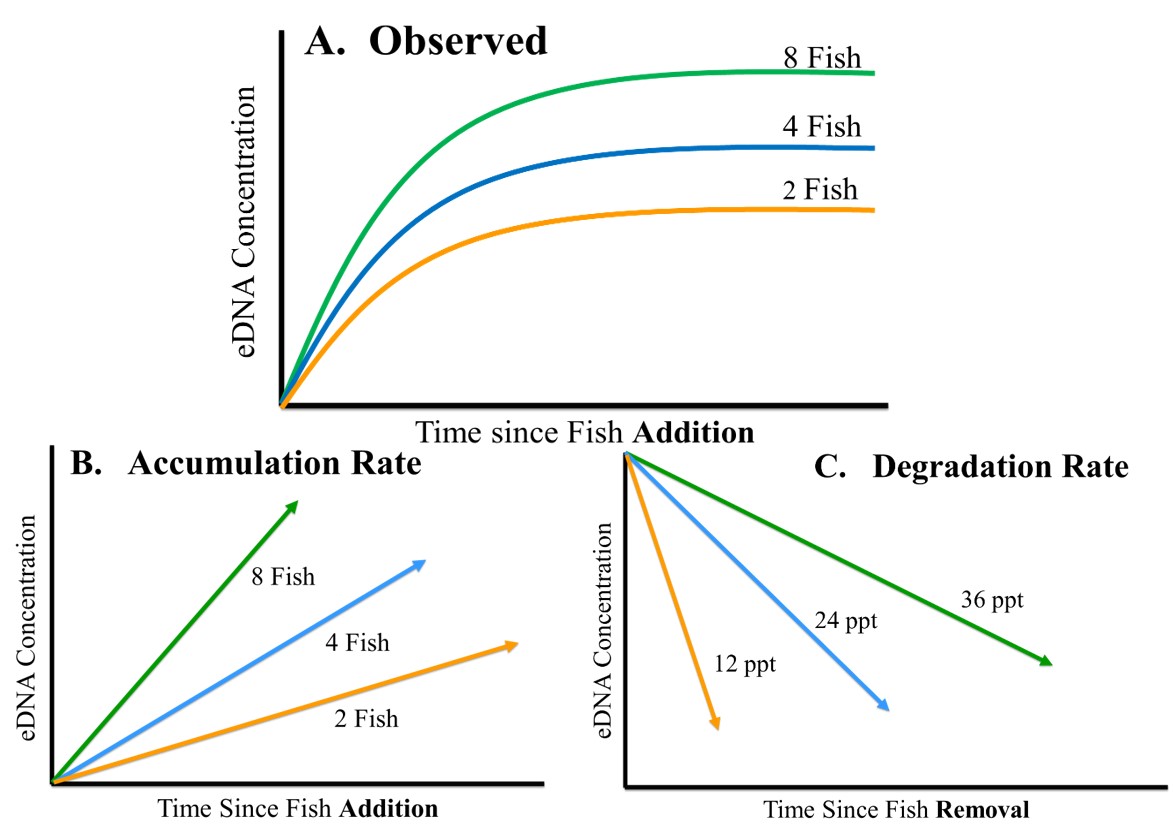
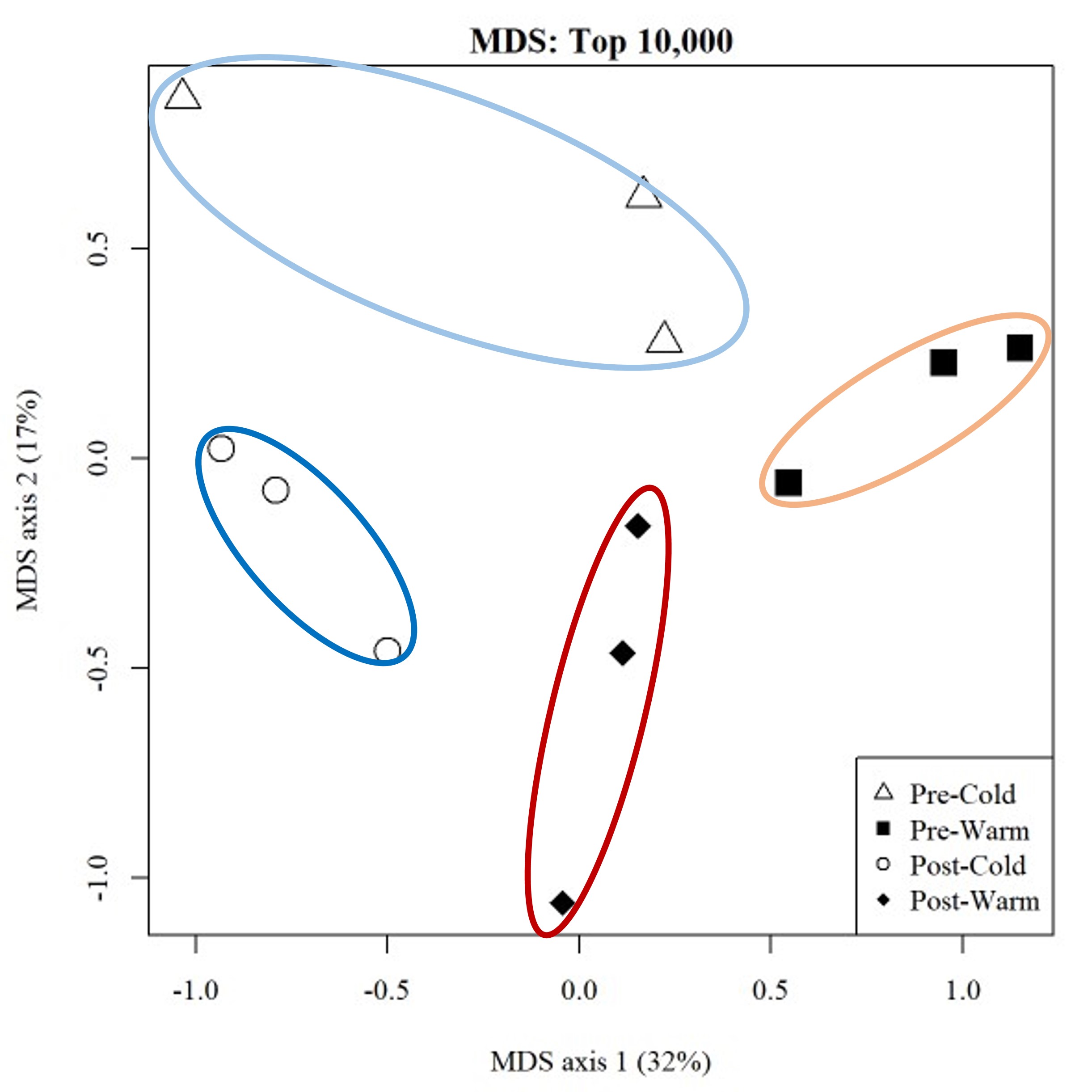
Understanding how genomic variation scales to phenotypic variation, on which natural selection acts, is a central theme in evolutionary biology. Using the northern two-lined salamander (Eurycea bislineata) and the spotted lanternfly (Lycorma delicatula), I am currently using whole genome sequencing along with physiological and morphometric data to explicitly link genetic variants with traits potentially under selection in urban landscapes. However, a full understanding of genome-phenome relationship requires interrogating the intermediate steps between genomic change and phenotypic changes (i.e., transcription & translation). Hence, I employ transcriptomic methods like RNAseq to understand how gene expression regulates phenotypic change. I have been particularly interested in plastic changes in gene expression driven by both genotypic and environmental variation. I have been exploring the GxE interaction both in relation to developmental temperature in the mangrove rivulus fish (Kryptolebias marmoratus) and paternal predator exposure in three-spined stickleback (Gasterosteus aculeatus) with plan to extend this to the spotted lanternfly.